Aperçu
NEMO¶
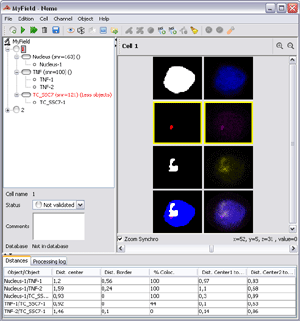
Three-dimensional fluorescence in situ hybridization (3D-FISH) is used to study the organization and the positioning of chromosomes or specific sequences such as genes or RNA in cell nuclei. Many different programs (commercial or free) allow image analysis for 3D-FISH experiments. One of the more efficient open-source programs for automatically processing 3D-FISH microscopy images is Smart 3D-FISH, an ImageJ plug-in designed to automatically analyze distances between genes. One of the drawbacks of Smart 3D-FISH is that it has a rather basic user interface and produces its results in various text and image files thus making the data post-processing step time-consuming. We developed a new Smart 3D-FISH graphical user interface, NEMO, which provides all information in the same place so that results can be checked and validated efficiently. NEMO give users the ability to drive their experiments analysis in either automatic, semi-automatic or manual detection mode. We also tuned Smart 3D-FISH to better analyze chromosome territories.
Download Nemo 1.5.1 for Windows 32 bits : https://forge-dga.jouy.inra.fr/attachments/download/6031/Nemo1.5.1-Win32.zip
Download Nemo 1.5 for Linux 32/64 bits : https://forge-dga.jouy.inra.fr/attachments/download/6032/Nemo1.5-Linux32.tar.gz
Download Nemo 1.5.2 patch (nemo.jar file) : https://forge-dga.jouy.inra.fr/attachments/download/6033/nemo.jar
Download Nemo user guide : https://forge-dga.jouy.inra.fr/attachments/download/593/NemoUserGuide1.1.pdf
Download Image Samples : https://forge-dga.jouy.inra.fr/attachments/download/6035/NemoImageSample.zip
Please cite this software using:
Iannuccelli E, Mompart F, Gellin J, Lahbib-Mansais Y, Yerle M, Boudier T: NEMO: a tool for analyzing gene and chromosome territory distributions from 3D-FISH experiments. Bioinformatics, 26(5):696-697.
Membres
Manager : Eddie Iannuccelli